Gut Microbiota and Its Research Tools
Scientists have discovered the correlation between intestinal flora and human diseases. In addition to helping develop new disease diagnosis and prognosis tools, it also provides a new way of thinking and approach to explore pathogenic mechanisms, including using changes in the microbiome as a new means of treating diseases. However, changes in intestinal flora are affected by the host's own genes as well as the environment, diet, lifestyle and disease status. The relationship between intestinal flora and disease involves a collection of many complex variables, and correlation studies alone cannot clarify the causal relationship between intestinal flora and disease. Therefore, the study of the correlation between intestinal microbiota and human diseases is only the beginning of intestinal microbiota research. Other research tools are still needed to establish the causal relationship and related mechanisms between intestinal bacteria and diseases, which will help the subsequent application and development of translational medicine. (Figure 1).
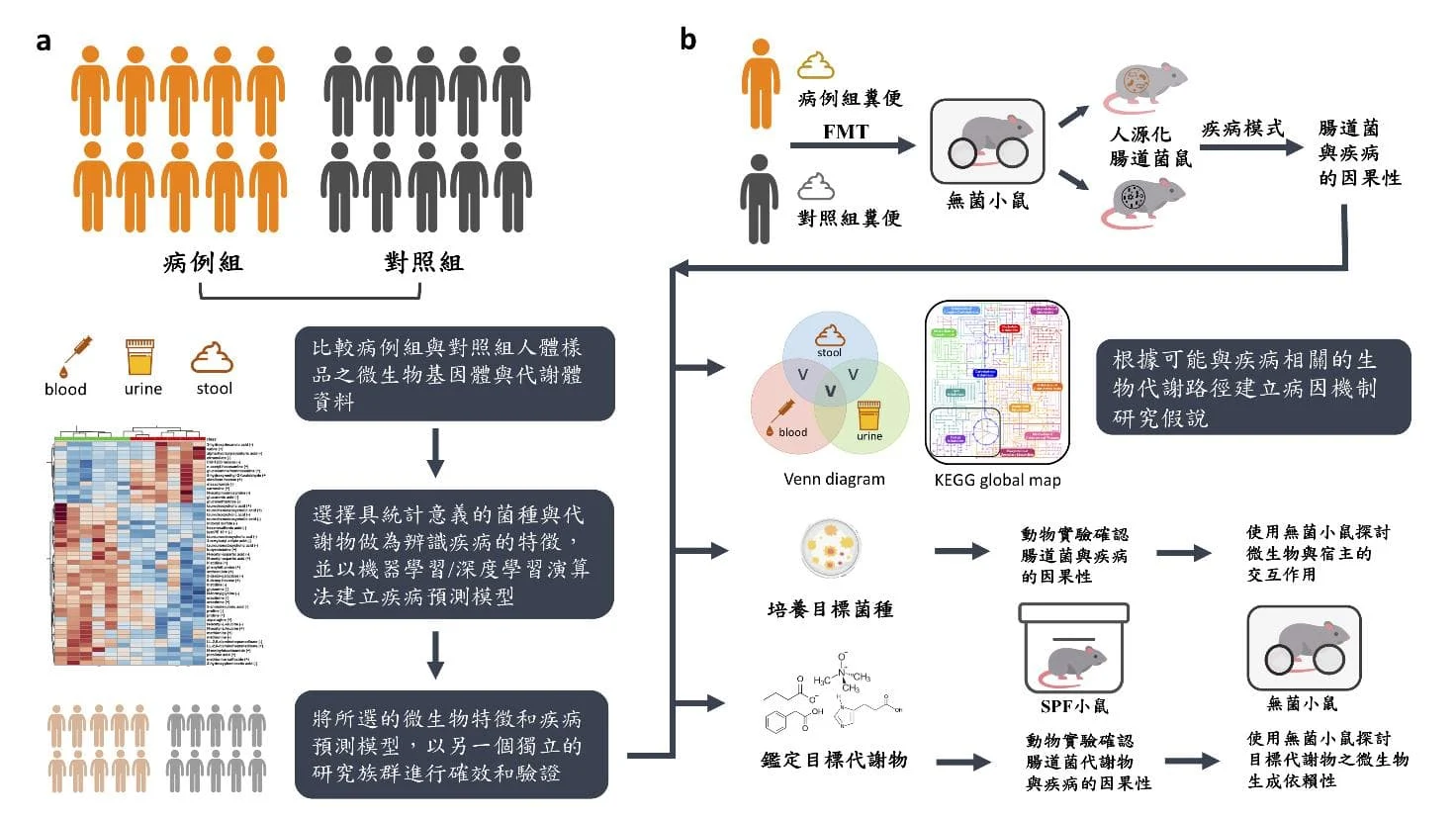
Figure 1: Methods and model tools for translational medicine research on the intestinal microbiota.
(a) Use case-control studies to identify disease-associated gut bacterial species and metabolites.
(b) Humanized intestinal bacteria mice were established using fecal microbiota transplantation to explore the causal relationship between intestinal bacteria and diseases. (modified from Wu WK et al. Rapid Commun Mass Spectrom 2020)
Microbial metagenomics
With the advancement of gene sequencing technology, intestinal microbiome studies have also flourished in the post-genomic era of humans. Many chronic diseases have been found to be highly correlated with specific intestinal microbial flora composition or functional genes, such as cardiovascular disease, diabetes, obesity, fatty liver, inflammatory bowel disease, rheumatoid arthritis, Alzheimer's disease, Parkinson's disease, autism, etc. Currently, common high-throughput sequencing equipment includes the short-read Illumina platform and the long-read PacBio and Nanopore platforms. Due to the advancement of instruments and reagents, the cost of sequencing has been greatly reduced. However, a large amount of sequence data still requires bioinformatics experts to conduct good quality control, classification, optimization and statistical analysis of the data.
Microbial metabolomics
Since DNA cannot represent the functional RNA and protein expression and post-translational modification in the association between intestinal bacterial species composition, microbial gene abundance and diseases, there is still a long way to go before using microbial genes to explain the mechanism of human diseases. In addition, many intestinal anaerobic bacteria are difficult to culture, the gene database is relatively incomplete, and the functions of many gene products have not been verified, making it easy for the interpretation of the association between genes and diseases to be biased. Therefore, directly detecting and analyzing intestinal bacterial metabolites has become one of the solutions to directly explore the functionality of intestinal bacteria. In recent years, scientists have discovered many intestinal bacterial metabolites that are biologically active and can regulate host physiology, including TMAO, imidazole propionate, phenylacetate, indole propionate, etc. They all use high-resolution mass spectrometry to analyze metabolites and compare them with diseases to identify candidate metabolites related to diseases, and then conduct in vitro and animal experiments to verify them. Therefore, for functional small molecules, metabolome analysis provides a relatively direct research platform for exploring active molecules of intestinal bacteria, which helps to accelerate the development of new intestinal bacteria small molecule drugs.
Germ-free mice and gnotobiotic mice model
After finding the correlation between intestinal microflora and disease, if we want to further explore the impact of intestinal bacteria on disease occurrence or treatment prognosis, we need to use appropriate animal experimental models to verify causality. Among them, the germ-free mice and gnotobiotic mice models are the gold standard for verifying the causal relationship between bacteria and diseases. However, the establishment and maintenance of sterile mouse facilities requires considerable manpower and technical costs. Not only does each isolator for breeding sterile mice need an independent air flow filtration system, but also special transportation equipment and sterilization procedures. The breeding, feeding, operation and quality control processes are complicated and difficult to simplify. Although this technology is difficult to maintain and has a high threshold for operation, it can not only compare whether the host phenotype or gene expression is affected by intestinal bacteria in the presence and absence of bacteria, but also colonize specific strains (mono-colonization) or specific bacterial communities (consortia colonization) in germ-free mice to study the interaction, causal relationship and molecular mechanism between microbes and hosts. It can also reveal how intestinal bacteria regulate host immunity, metabolism and neural function, or the context of bacterial imbalance leading to human diseases, and provide a deeper understanding. In terms of translational medicine applications, human feces with different disease phenotypes can also be transplanted into the intestines of germ-free mice to establish a humanized gnotobiotic mice (or human microbiota associated mice) model to verify the results observed in clinical microbial genome sequencing analysis to see whether there is a causal relationship between microbiota and disease phenotypes.
Anaerobic culturomics
The development of human intestinal microbial research has been unable to be satisfied with the discovery of the correlation between microbial flora and diseases. Even if specific bacterial species that may cause or protect against diseases are found through bacterial genome sequencing analysis, the target bacterial species still needs to be subjected to in vitro and animal experiments to verify the research hypothesis. Only by isolating and culturing the target strain can a more clear pathogenic mechanism be explored for the specific bacterial species. Since the human intestine is an anaerobic environment, most of the bacteria in the intestine are anaerobic bacteria, or even absolute anaerobic bacteria that are extremely sensitive to oxygen. Once they come into contact with the outside air, they quickly lose their activity and cannot be activated and cultured. In addition, due to the large number and variety of microorganisms in the human intestine, the number of bacteria in every gram of feces is as high as 1011~1012, and the number of species is as high as thousands. Some bacteria with low relative abundance (relative abundance ≈ 0.01%) are difficult to select and separate from thousands of strains on the culture medium even if they can be cultured in vitro. Therefore, anaerobic bacterial culture has always been a difficult problem in studying intestinal bacteria in the past. However, in recent years, with the advancement of anaerobic culture and bacterial species identification methods, as well as the establishment of a human microbial genome database, many intestinal bacteria that were previously thought to be uncultivable have gradually been cultured, isolated, and identified. In particular, the microbial culture team led by Didier Raoult of the University of Marseille in France used a variety of culture conditions and a self-built MALDI-TOF database for rapid bacterial species identification. In just a few years, they were able to culture most of the previously known intestinal microbes, and further isolate and culture more than 500 new strains of human intestinal bacteria. This pioneering effort has opened up a ray of hope for the anaerobic culture of intestinal bacteria, which was difficult to achieve in the past. It is expected that more important microbes related to human diseases will be discovered, cultured, and their significance will be elucidated in the future, creating new opportunities for precision medicine for humans.
in conclusion
The intestinal microbiome is a biomedical field that has emerged in recent years. Like artificial intelligence, it is believed to have the potential to bring about tremendous changes in modern medicine. Currently, related research is in the embryonic stage of rapid development. Although it has not yet fully blossomed in clinical applications, it has already brought about many new breakthroughs in thinking. In the future, the development of intestinal microbes will move from the discovery of associations to causal research, and finally return to cultivation to explore the mechanism of action between microbes and the host, placing the last piece of the puzzle towards the goal of precision medicine.



